For Cas9-triggered homologous recombination you need two constructs:
- A Cas9-sgRNA expression plasmid
- A homologous repair template plasmid
Cas9-sgRNA expression plasmid
The Cas9-sgRNA plasmid is generated by choosing the 20 bp sgRNA target sequence, then inserting it into an expression construct using site-directed mutagenesis. Detailed instructions for target site selection and cloning are in the protocol.
Several different Cas9-sgRNA expression constructs are available from Addgene:
- pDD162 is our original construct for co-expression of both Cas9 and sgRNA. sgRNA expression is driven by the R07E5.16 U6 promoter.
- pJW1219 is a pDD162 derivative with the modified "F+E" sgRNA scaffold. It has been reported to have higher activity.
- pDD121 expresses Cas9 only (without any associated sgRNA) for use when the sgRNA is expressed from another plasmid (for example, the combined sgRNA + repair template constructs that can be created with SapTrap cloning).
- Addgene also has several other plasmids for Cas9 and sgRNA expression in C. elegans, but we have not tested these.
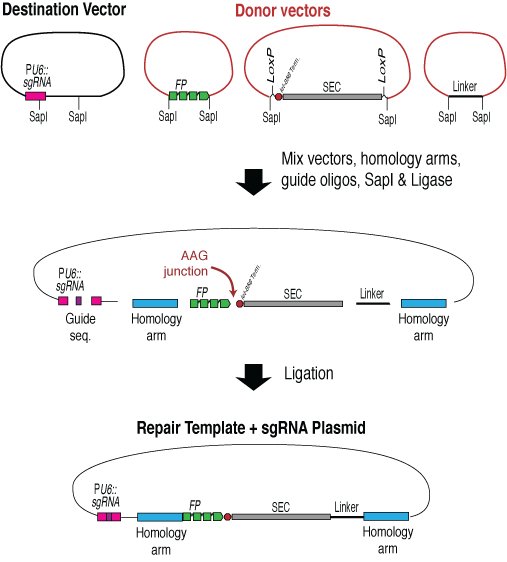
Homologous repair template - SapTrap cloning method
Matt Schwartz in Erik Jorgensen's lab has developed a modular assembly for CRISPR repair templates, referred to as "SapTrap" cloning. This approach is nice because it allows a user to mix and match different functional parts and assemble a repair template in a modular fashion. The basic workflow is:
- Obtain the homology arms, either by PCR or as synthetic DNA fragments. Optionally, purchase oligos with the sequence of the sgRNA target if you want to also include the sgRNA in the repair template construct.
- Mix the homology arms, donor vectors carrying an FP, SEC and any desired linkers, optionally the sgRNA oligos, and a destination vector in a SapTrap assembly reaction. Transform the reaction into E. coli.
- Isolate DNA from a few clones and verify by restriction digestion and sequencing.
SapTrap plasmid kits from us and from the Jorgensen lab are available from Addgene
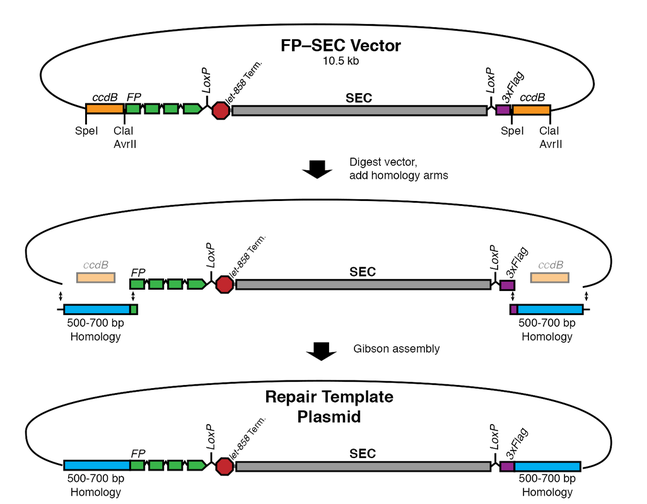
Homologous repair template - ccdB-based cloning method
In our 2015 protocol, the homologous repair template is generated by cloning homology arms into an FP–SEC vector. Although we now mostly use SapTrap instead, this approach still works well and is useful in certain situations - for example, when the homology arms contain SapI sites.
The procedure involves four simple steps:
The procedure involves four simple steps:
- Digest the vector with restriction enzymes to remove the ccdB negative selection markers. ccdB is toxic to E. coli, and is used to facilitate homology arm insertion. A batch of digested vector can be used to make many repair templates.
- Amplify the homology arms by PCR, using primers that add sequence overlaps for Gibson assembly. Alternatively, purchase the homology arms as synthetic DNA fragments.
- Mix the homology arms and digested vector in a Gibson assembly reaction. Transform the reaction into E. coli. Because of the ccdB negative selection, only clones that have correctly inserted the homology arms will grow.
- Isolate DNA from a few clones and sequence to verify homology arm insertion. The procedure is so efficient that screening clones before sequencing is not necessary.
The following FP–SEC vectors are available from Addgene:
Want to make an FP–SEC vector with your favorite tag? Feel free to contact us for cloning suggestions. We encourage everyone to share their FP–SEC vectors with the community – please deposit them at Addgene, and let us know so that we can link to them from this page.
- mTagBFP^SEC^3xFlag (pUA97). mTagBFP is a blue fluorescent protein derived from the same scaffold as TagRFP. This construct was made and deposited at Addgene by Oliver Hobert's lab.
- mTagBFP2^SEC^3xFlag (pJJR81). mTagBFP2 is a blue fluorescent protein derived from the same scaffold as TagRFP. This construct was made and deposited at Addgene by Mike Boxem's lab.
- mTurquoise2^SEC^2xHA (pDD315 - annotated ApE file). mTurquoise2 is a cyan fluorescent protein derived from GFP.
- GFP^SEC^3xFlag (pDD282 - annotated ApE file). The GFP in this vector is codon-optimized for C. elegans and has the S65C mutation found in the original GFP vectors from the Fire lab.
- EGFP^SEC^3xFlag (pJJR82). The GFP in this vector is codon-optimized for C. elegans and has the S65T mutation found in the EGFP constructs that are commonly used in other organisms.
- YPET^SEC^3xFlag (pDD283 - annotated ApE file). YPET is a brighter but less photostable derivative of YFP.
- TagRFP-T^SEC^3xMyc (pDD286 - annotated ApE file). TagRFP-T is an orange-red fluorescent protein that is spectrally similar to DsRed and other old-school RFPs. This construct has Lox2272 sites in place of LoxP, allowing it to be used in a background that has already been modified with one of our green vectors.
- mCherry^SEC^3xFlag (pJJR83). This construct was made and deposited at Addgene by Mike Boxem's lab.
- mKate2^SEC^3xMyc (pDD287 - annotated ApE file). mKate2 is a red fluorescent protein that is spectrally similar to mCherry. This construct has Lox2272 sites in place of LoxP, allowing it to be used in a background that has already been modified with one of our green vectors.
- Pmyo-2::GFP + SEC (pDD317 - annotated ApE file). This vector is intended for knocking out a gene and replacing it with Pmyo-2::GFP.
- TagRFP-T^SEC^3xFlag (pDD284 - annotated ApE file). Note: We now consider this construct obsolete and recommend pDD286 instead.
- mKate2^SEC^3xFlag (pDD285 - annotated ApE file). Note: We now consider this construct obsolete and recommend pDD287 instead.
Want to make an FP–SEC vector with your favorite tag? Feel free to contact us for cloning suggestions. We encourage everyone to share their FP–SEC vectors with the community – please deposit them at Addgene, and let us know so that we can link to them from this page.
Our old construct design page is available here.